Lecturers
- Nenad Ban, ETH, Zürich, Switzerland: Protein structure& function; X-ray diffraction
- Ribosomes and their functional complexes
- Beyond the prokaryotic ribosome
- Eurico Cabrita, Universidade Nova de Lisboa, Portugal
- Molecular Recognition, Intermolecular and Protein ligand interactions, NMR
- Mario Cindrić, Ruđer Bošković Institute
- Mass Spectrometry for protein sequencing
- Edwin De Pauw, Uni Liége, Belgium
- Mass spectrometry of nucleic acids structures and complexes
- Helena Danielson, Beactica AB, Uppsala, Sweden
- QCMD/SPR
- Zvonimir Dogić, Brandeis, USA
- Complex fluids, self-assembly
- Marija Drndić, University of Pennsylvania, USA: Experimental nano/biophysics
- biomolecular analysis with nanopores
- Peter Kolb, Philipps-Universität Marburg, Germany
- Chemoinformatics, docking to GPCRs
- Sabrina Leslie, McGill University, Montreal, Canada
- nanoconfinement of DNA
- Fraser MacMillan, University of East Anglia, UK
- EPR spectroscopy
- Chris Oostenbrink, UNRLF, Vienna, Austria: Structure and dynamics of biomolecules; modeling
- Ensembles and sampling, leading to molecular dynamics simulations
- Structure refinement using molecular dynamics simulations (NMR observables)
- Calculation of free energies from molecular simulation
- Arwen Pearson, Uni Hamburg, Germany: time-resolved X-ray and SAXS
- data collection methodologies, library of photocaging chemistries
- complementary solution phase experiments, THz time-domain spectroscopy
- Ana-Sunčana Smith, Ruđer Bošković Institute & Uni Erlangen-Nürnberg Germany: Membranes and vesicles, cell adhesion
- Membranes, from self assembly to rafts 1,2,3
- Frank Sobott, Uni Antwerpen, Belgium
- “native” mass spectrometry, ion mobility: structural proteomics techniques
- Holger Stark, MPI-BPC, Göttingen, Germany: Protein structure& function; cryoEM
- Introduction into single particle cryo-EM
- High-resolution structure determination of macromolecular complexes by cryo-EM
- Antonio Šiber, Institute of physics, Zagreb, Croatia: Physics of viruses
- Physics of viruses: electrostatics, elasticity and DNA condensation in viruses 1,2,3
- Iva Tolić, Ruđer Bošković Institute, Zagreb: Physics of cells
- Forces in the mitotic spindle
- Live-cell fluorescence microscopy and laser-cutting inside the cell
- Nuška Tschammer, Nanotemper, Germany
- GPCR protein-ligand recognition
- Dmitry Veprintsev, PSI, Villigen, Switzerland: Protein structure&function; X-rays & NMR
- Conformational states and ligand-induced changes in GPCRs
- Meni Wanunu, Northeastern Uni, USA
- Nanoscale biophysics, nanopores
- Anthony Watts, Department of Biochemistry, Oxford, UK: Protein structure& function; solid state NMR
- Principles of solid state NMR for the study of biomolecules
- Solid state NMR for structural studies of large integral membrane proteins
- Receptor dynamics and structure in membranes resolved using solid state NMR
- Bojan Žagrović, MFPL, Vienna, Austria: Computational Biophysics of Macromolecules
- More dynamic than we think? On conformational averaging in structural biology 1, 2
- Protein-RNA interactions and the origin of the genetic code
Download final program: Program_13th-International-School-of-Biophysics
Short-talks (8+2 minutes) by students:
1.9.2016 Protein machines
Shiran Barber Zucker: Hypermanganesemia as a Result of a Cation Diffusion Facilitator Cytoplasmic Domain Loss of Structure
Jelena Dragojević: Functional characterization of organic anion transporters in zebrafish (Danio rerio): Oat2a-d
Franziska Heydenreich: A GPCR as a Michalis-Menten enzyme: Understanding biased signalling of vasopressin V2 receptor
Faezeh Mofidi Najjar: Mechanistic study on catalase activation by curcumin
Stephan Rempel: The new vitamin B12 degradation product transporter BtuM
Tomas Šneideris: Looking for a generic inhibitor of amyloid-like fibril formation among flavone derivatives
Olga Tkachenko: Chaperone Mechanism of αB-Crysrallin
Oleksii Zdorevskyi: Understanding the mechanism of DNA deactivation in ion therapy of cancer cells: hydrogen peroxide action
2.9.2016 MS, EPR, X-ray, NMR
Miranda Collier: Dynamic small heat-shock protein interactions with non-aggregating clients
Christoph Gmeiner: Pulse EPR with Gd(III)-spin labels to determine distance distributions in a large protein/RNA complex
Daniel Mayer: The structural basis for recognition of the „Phosphorylation Barcode“ of G protein-coupled receptors by arrestin
Anna Mullen: Understanding structures & functional dynamics of membrane proteins using EPR spectroscopy
Marija Nišavić: Positive/negative ion mode nano-electrospray ionization mass spectrometry of metallated peptides
Agnieszka Sztyler: Analysis of intrinsically disordered region of human topoisomerase I using hydrogen/deuterium exchange mass spectrometry
Lara Štajner: Investigation of selected amino acids influence on calcium carbonate precipitation as simple models of organic matrix
4.9.2016 MD/simulations/models
Andrea Cesari: Using the maximum entropy principle to enforce NMR data on RNA simulations
Yossa Dwi Hartono: Investigation of tautomerisation and protonation equilibria of nucleic acid with lambda dynamics
Willem Jespers: Binding mode prediction using free energy perturbations
Jakub Kollár: Computational design of selective histone deacetylase inhibitors as epigenetic agents targeting cancer
Negar Nahali: Density effects in entangled solutions of ring polymers
5.9.2016 Lipid Membrane/ Membrane proteins
Barbara Eicher: Joint analysis of SAXS and SANS data of asymmetric lipid vesicles
Dietmar Hammerschmid: Characterization of the globin coupled sensor from Geobacter sulfurreducens using native mass spectrometry
Steven Lavington: Exploring NTS1 helix 8 conformation and dynamics in lipid environments
A. Skarabahatava: In vitro and in vivo changes in lipids erythrocyte membrane affected aluminum ions
Lilit Tonoyan: The Influence of Sodium Dodecyl Sulfate on Stability of Bilayer Lipid Membranes
Maria Tsemperoulli: Optical and Electrical Characterization of Voltage Sensitive Dyes in Lipid Bilayers
6.9.2016 Optics & imaging based approaches
Alexandra Falamas: Raman microspectroscopy of stem cells: approach to monitor the CBCT radiation effect
Barbora Kalouskova: Preparation and Structural Characterization of Human NK Cell Activating Immunocomplex NKp80:ACIL
Ana Milas: Sliding of microtubules in the bridging fiber segregates chromosomes by creating poleward flux of the attached k-fibers
Rita Nagypál: Fluorescence kinetics of flavin adenine dinucleotide in different microenvironments
Aiswaria Prakash: Position dependent fluorescent properties of coupled fluorescent dyes in RNAs and proteins
Ioana Maria Simon: Study of the biological effects of oral diagnostic irradiation with Raman spectroscopy
Dora Sviben: Stability of proteins and viruses determined by differential scanning fluorimetry
8.9.2016 Nanobio/soft matter physics
Jiandong Feng: Coulomb blockade in ion transport
Sanjin Marion: Nucleic acids and condensing proteins in viral confinement
Martina Požar: The microscopic structure of hot and cold alcohol mixtures
Biljana Stojković: Micro and macro-rheology study of DNA-levan mixtures
Pradeep Waduge: Angstrom-Sensitivity Protein Sizing and Conformation Detection using Solid-State Nanopores
NanoTemper’s MST and nanoDSF Workshop
The workshop will cover principles of biomolecular interaction analytics using Microscale Thermophoresis (MST) as well as protein stability and aggregation assessment with use of nanoDSF technology. We strongly encourage to bring your samples for analysis. Participants interested in measurements of binding affinities or protein stability of their own samples are requested to contact Dr. Marek Zurawski ([email protected]) for further information.
Number of participants: 32 (working in pairs). One student pair will have about 4 hours of exp.work with NanoTemper equipment. Experiment slots:
Sep1: 15-19h; Sep2: 9-13h, Sep2: 15-19h; Sep3: 8-12h
Molecular dynamics simulations
This workshop will help you gain first experiences in performing molecular dynamics simulations. After a short theoretical introduction, you will find out about the basic ingredients for a molecular simulation. You will setup your own simulation of a small peptide and run some short simulations of it. Finally, you will analyse the resulting trajectories and find out what computational methods can offer in addition to experimental approaches.
Number of participants: 20. Students will be introduced to their task by Prof. Bojan Zagrovic on Sep 1, from 16:00h, while Profs. Oostenbrink and Kolb will join on Sep 2nd and 3rd to wrap up. Work on Linux PCs at the Faculty of Science, or you bring your own Linux laptop.
Nanotechnology to the rescue: nanopores for DNA and protein sequencing
Biological and, in the last decade, solid-state nanopores (nm sized openings in nm thin membranes) are regarded as the most promising route towards the goal of reading-out individual genomes and even proteomes. The technique is based upon electrical read-out of the sequence while the biomacromolecule translocates through the nanopore. High-bandwidth (high-speed) preamps are the essential tool in achieving this, as well as the technologies to prepare and mount nanopores (prepared in e.g. graphene or MoS2) and control the translocation.
Number of participants: 20. Students will be split in groups of 5. Each group will be lead by one of the students who already work in this field, while Profs. Wanunu and Drndic will supervise. Work in labs to be set-up at the Faculty of Science.
Sequencing 2: Revenge of the chemists
Mass spectrometry is routinely used to identify pathogens and malignant cell strains by directly fingerprinting their proteome. Prof. Mario Cindirc has developed this method to the next level where it can be used for de-novo sequencing of peptides. The method is based on a proprietary chemical derivatization agent for preparation of the protein samples for MS analysis and heavily involves bioinformatics tools to analyze the MS spectra.
Number of participants: 12. Students will go to the Proteomics centre in Zagreb for a site visit and demonstration of the technology. Transfer by cars, accommodation 2 nights in Zagreb. Departure Sep 1. 17:00h, Return to Split Sep.3 before noon.
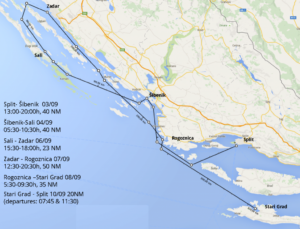
This year’s school takes place along the Adriatic coast on board cruise ships
Port of call: September 1-3: Split (Dalmatian Capital since Roman Times)
Programme & organization Committee
- Tomislav Vuletić, Institute of physics, Zagreb, Croatia (chair)
- Fraser MacMillan, CM1306 COST Action Coordinator, University of East Anglia, UK
- Peter Kolb, CM1207 COST Action Coordinator, Uni Marburg, Germany
- Frank Sobott, BM1403 COST Action Coordinator, Uni Antwerpen, Belgium
- Sanja Josef Golubić, University of Zagreb, Croatia
- Nadica Maltar-Strmečki, Ruđer Bošković Institute, Zagreb, Croatia
- Ida Delač Marion, Institute of Physics, Zagreb, Croatia
- Sanjin Marion, Institute of Physics, Zagreb, Croatia











